Accelerating New Drug Discovery
Introduction
Drug discovery, as the source of medicine innovation, is an important part of new medicine research and development. Drug discovery is implemented by target selection and confirmation. When fragments or lead compounds are discovered, similar compounds are usually searched in internal or commercial compound libraries in order to discover structure-activity relationship (SAR), compound availability, thus evaluating the potential of the lead compounds to be optimized to candidate compounds.
In order to discover available compounds in the fragment space from billion-scale compound libraries, chemical fingerprint is usually retrieved for substructure search and similarity search. However, the traditional solution is time-consuming and error-prone when it comes to billion-scale high-dimensional chemical fingerprints. Some potential compounds may also be lost in the process. This article discusses using Milvus, a similarity search engine for massive-scale vectors, with RDKit to build a system for high-performance chemical structure similarity search.
Compared with traditional methods, Milvus has faster search speed and broader coverage. By processing chemical fingerprints, Milvus can perform substructure search, similarity search, and exact search in chemical structure libraries in order to discover potentially available medicine.
System overview
The system uses RDKit to generate chemical fingerprints, and Milvus to perform chemical structure similarity search. Refer to https://github.com/milvus-io/bootcamp/tree/master/solutions/molecular_similarity_search to learn more about the system.
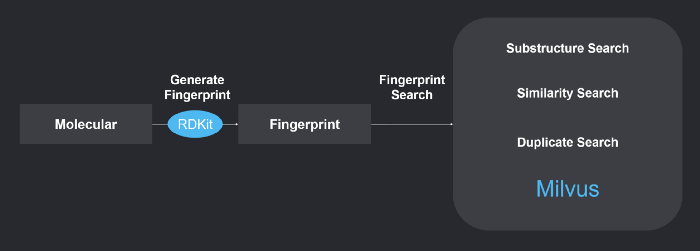 1-system-overview.png
1-system-overview.png
1. Generating chemical fingerprints
Chemical fingerprints are usually used for substructure search and similarity search. The following image shows a sequential list represented by bits. Each digit represents an element, atom pair, or functional groups. The chemical structure is C1C(=O)NCO1.
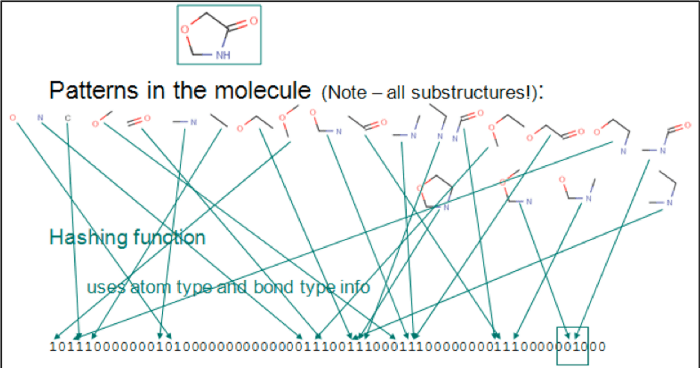 2-identifying-patterns-molecules.png
2-identifying-patterns-molecules.png
We can use RDKit to generate Morgan fingerprints, which defines a radius from a specific atom and calculates the number of chemical structures within the range of the radius to generate a chemical fingerprint. Specify different values for the radius and bits to acquire the chemical fingerprints of different chemical structures. The chemical structures are represented in SMILES format.
from rdkit import Chem
mols = Chem.MolFromSmiles(smiles)
mbfp = AllChem.GetMorganFingerprintAsBitVect(mols, radius=2, bits=512)
mvec = DataStructs.BitVectToFPSText(mbfp)
2. Searching chemical structures
We can then import the Morgan fingerprints into Milvus to build a chemical structure database. With different chemical fingerprints, Milvus can perform substructure search, similarity search, and exact search.
from milvus import Milvus
Milvus.add_vectors(table_name=MILVUS_TABLE, records=mvecs)
Milvus.search_vectors(table_name=MILVUS_TABLE, query_records=query_mvec, top_k=topk)
Substructure search
Checks whether a chemical structure contains another chemical structure.
Similarity search
Searches similar chemical structures. Tanimoto distance is used as the metric by default.
Exact search
Checks whether a specified chemical structure exists. This kind of search requires exact match.
Computing chemical fingerprints
Tanimoto distance is often used as a metric for chemical fingerprints. In Milvus, Jaccard distance corresponds with Tanimoto distance.
 3-computing-chem-fingerprings-table-1.png
3-computing-chem-fingerprings-table-1.png
Based on the previous parameters, chemical fingerprint computation can be described as:
 4-computing-chem-fingerprings-table-2.png
4-computing-chem-fingerprings-table-2.png
We can see that 1- Jaccard = Tanimoto. Here we use Jaccard in Milvus to compute the chemical fingerprint, which is actually consistent with Tanimoto distance.
System demo
To better demonstrate how the system works, we have built a demo that uses Milvus to search more than 90 million chemical fingerprints. The data used comes from ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/CURRENT-Full/SDF. The initial interface looks as follows:
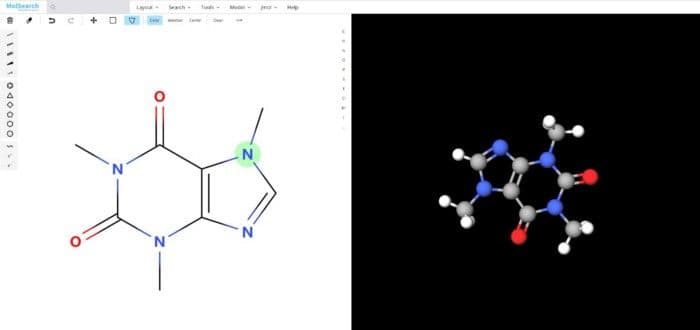 5-system-demo-1.jpg
5-system-demo-1.jpg
We can search specified chemical structures in the system and returns similar chemical structures:
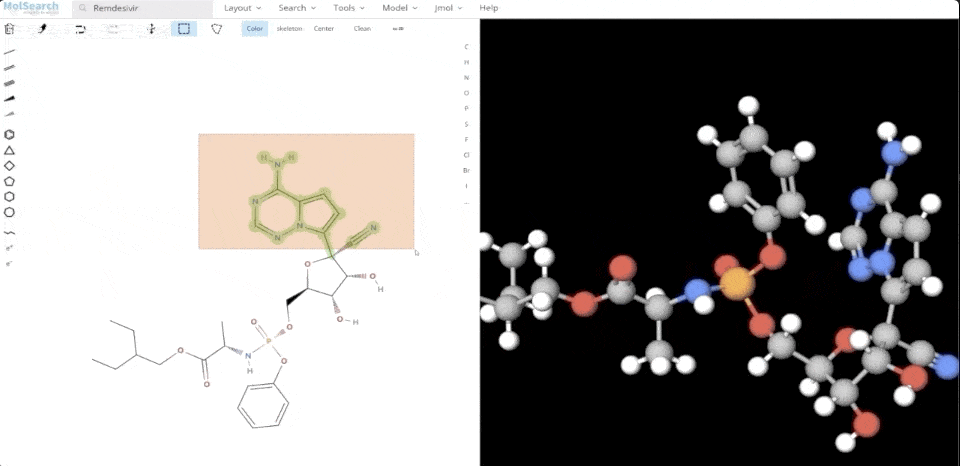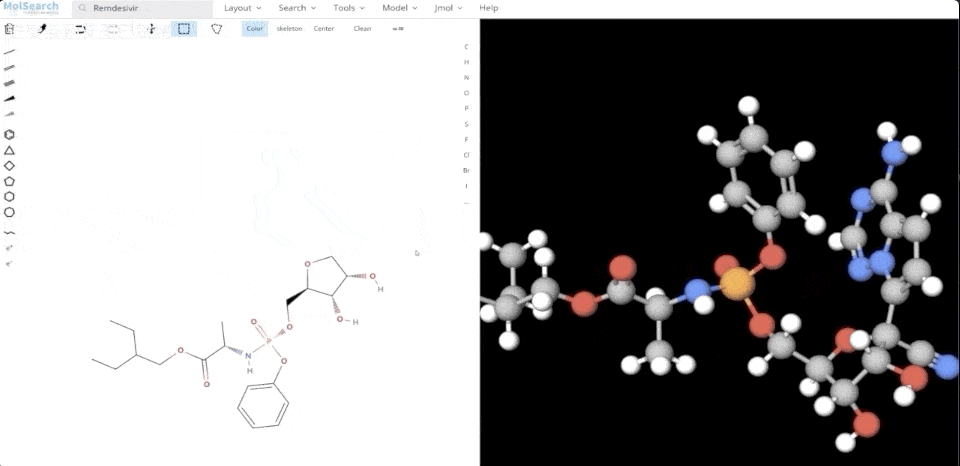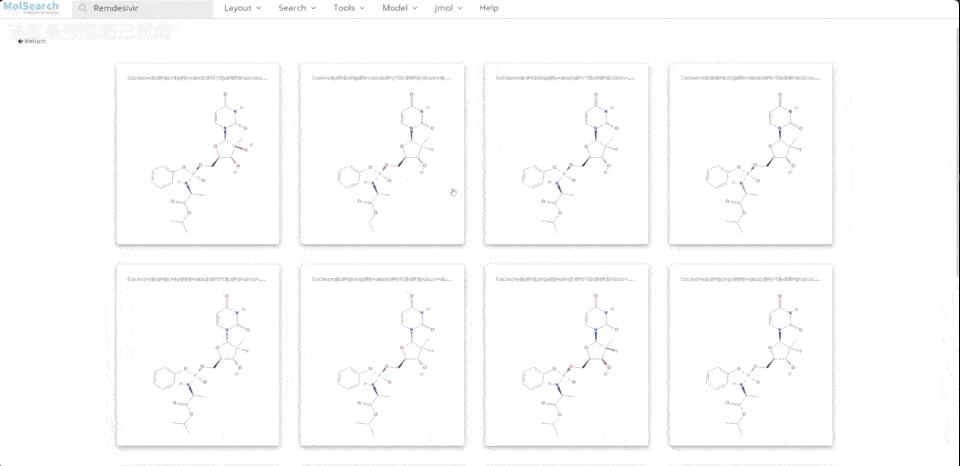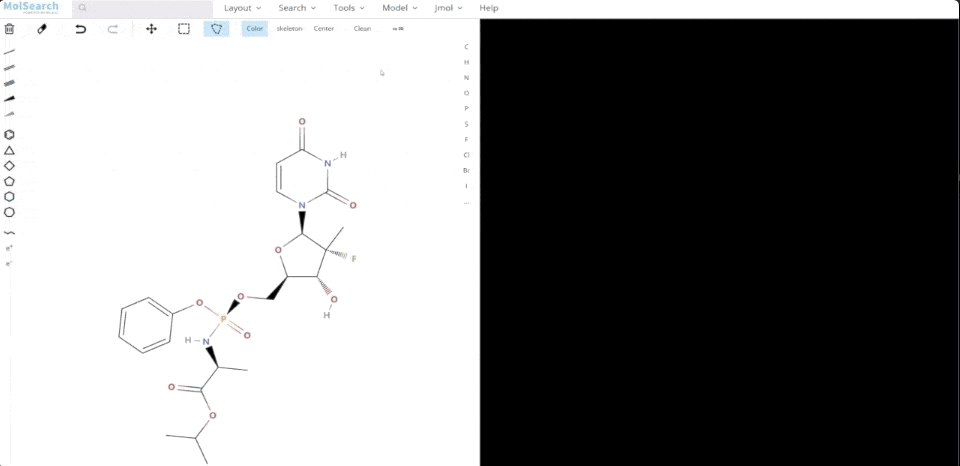 6-system-demo-2.gif
6-system-demo-2.gif
Conclusion
Similarity search is indispensable in a number of fields, such as images and videos. For drug discovery, similarity search can be applied to chemical structure databases to discover potentially available compounds, which are then converted to seeds for practical synthesis and point-of-care testing. Milvus, as an open-source similarity search engine for massive-scale feature vectors, is built with heterogeneous computing architecture for the best cost efficiency. Searches over billion-scale vectors take only milliseconds with minimum computing resources. Thus, Milvus can help implement accurate, fast chemical structure search in fields such as biology and chemistry.
You can access the demo by visiting http://40.117.75.127:8002/, and don’t forget to also pay a visit to our GitHub https://github.com/milvus-io/milvus to learn more!
Like the article? Spread the word